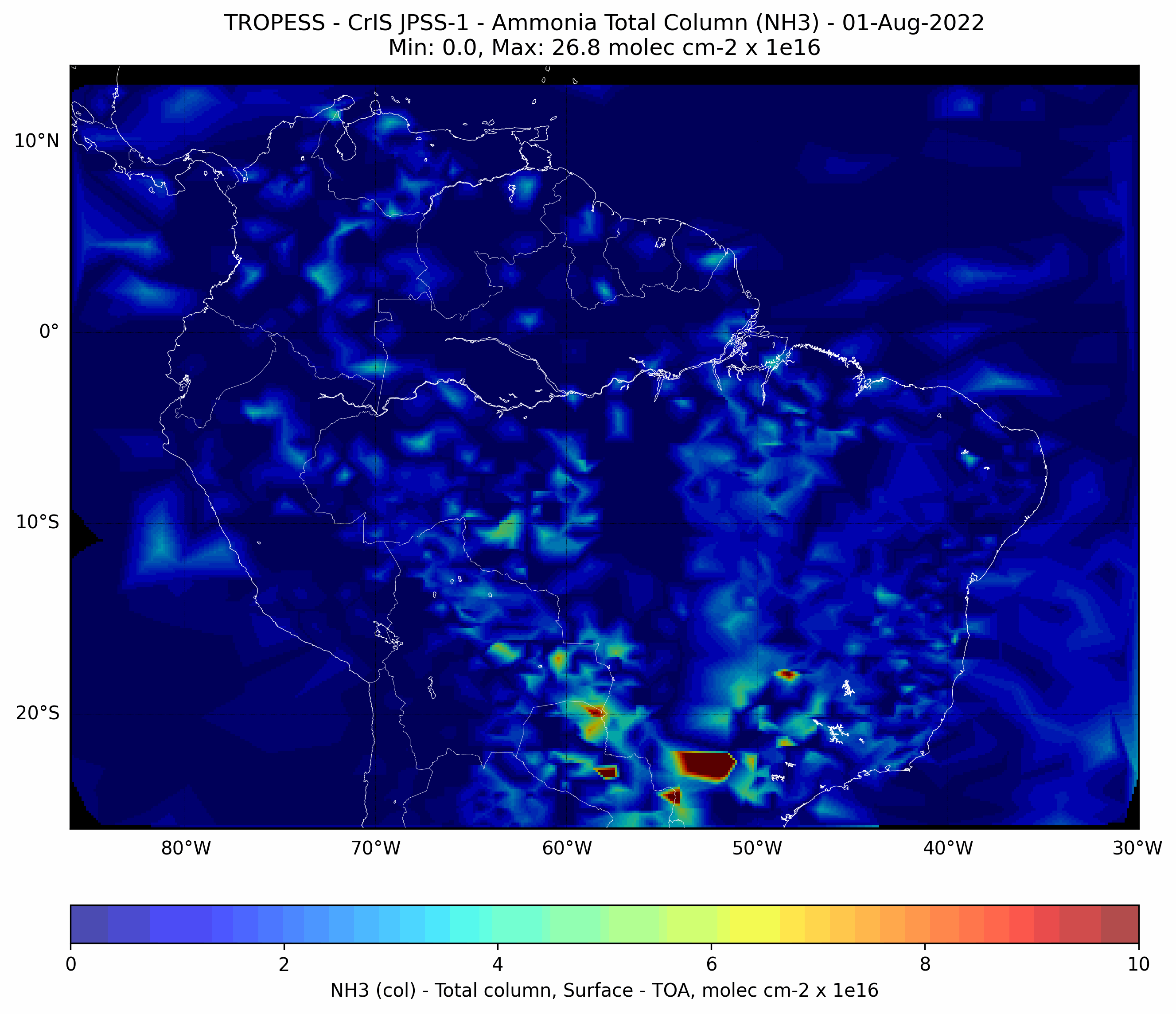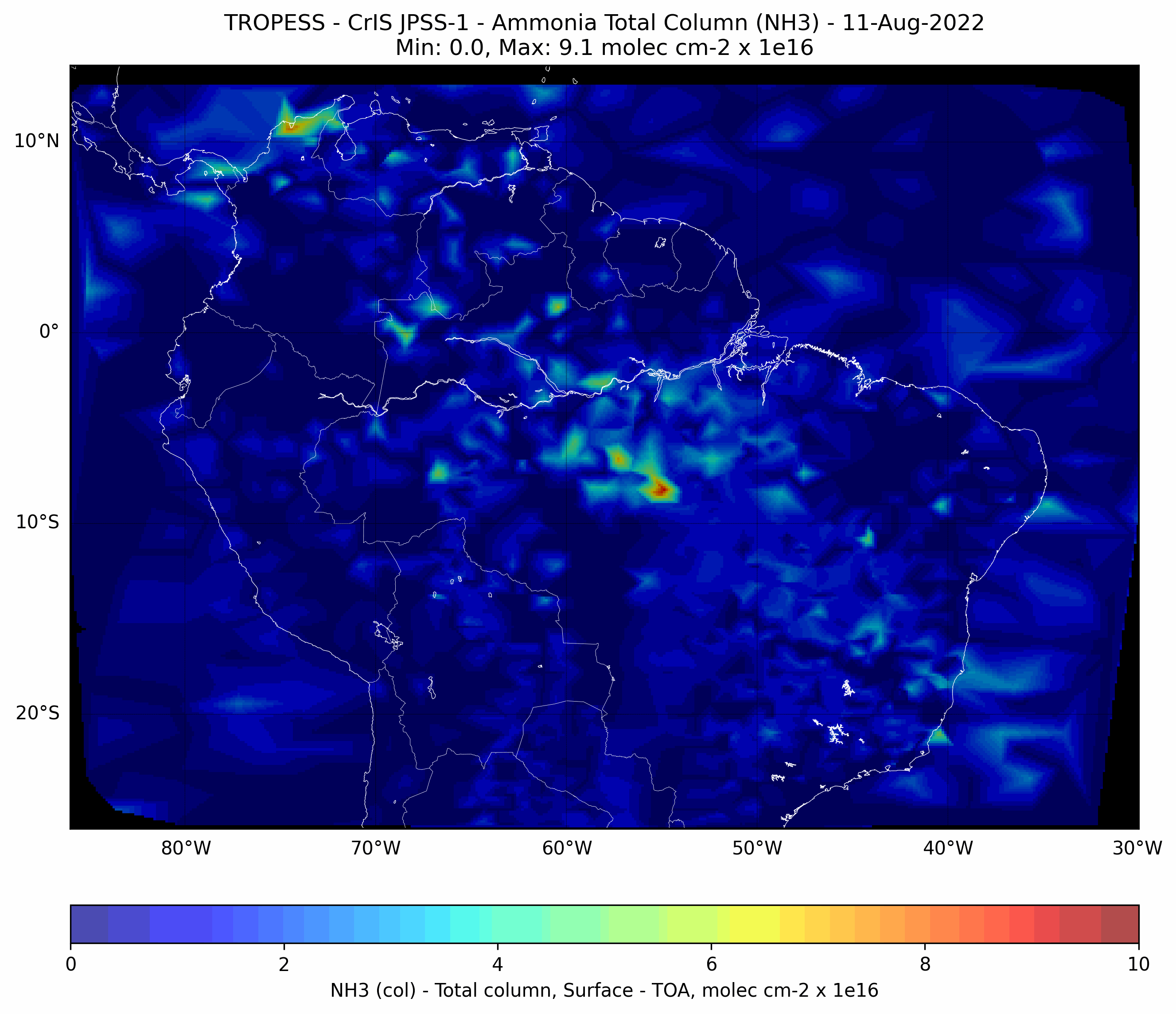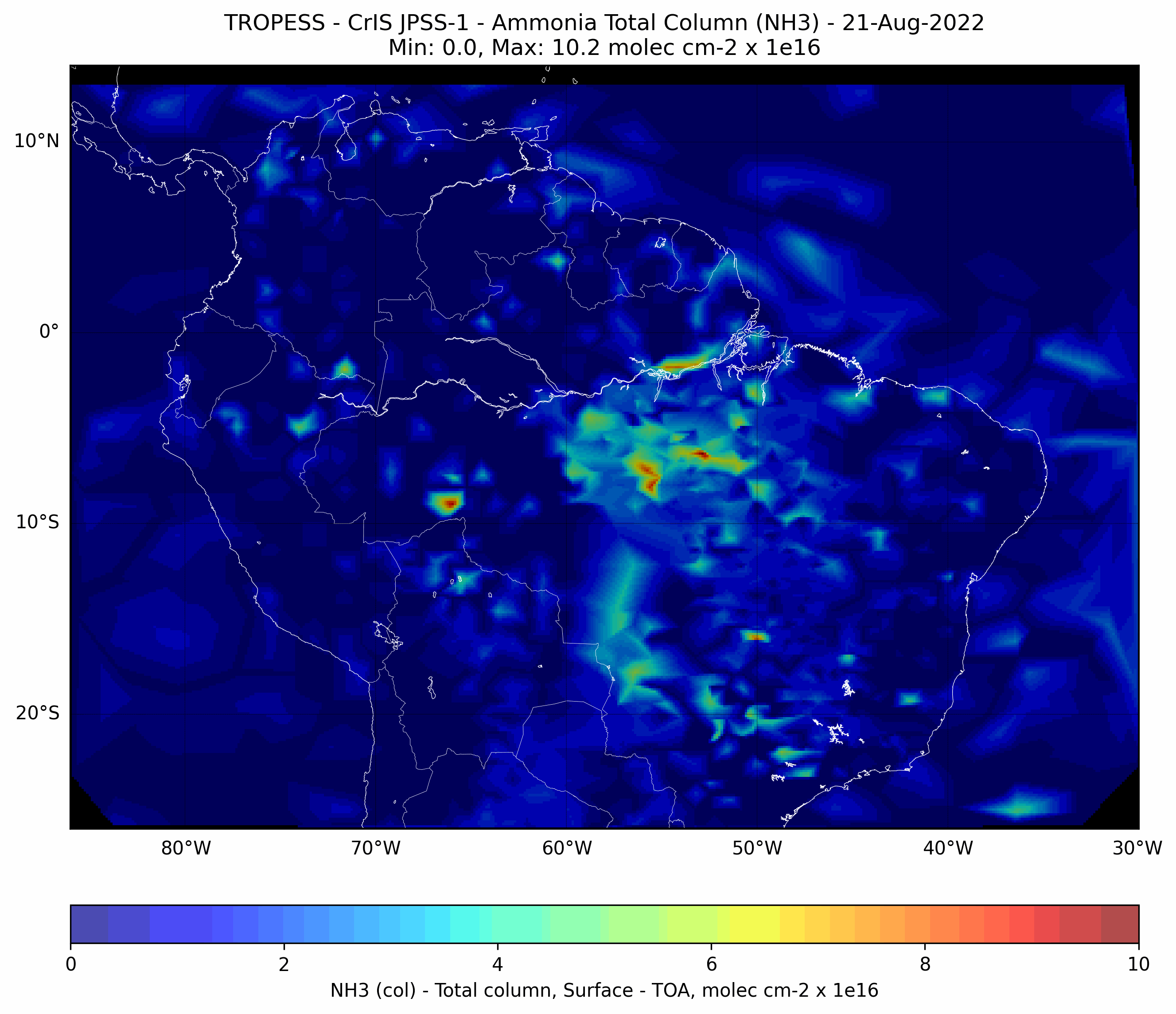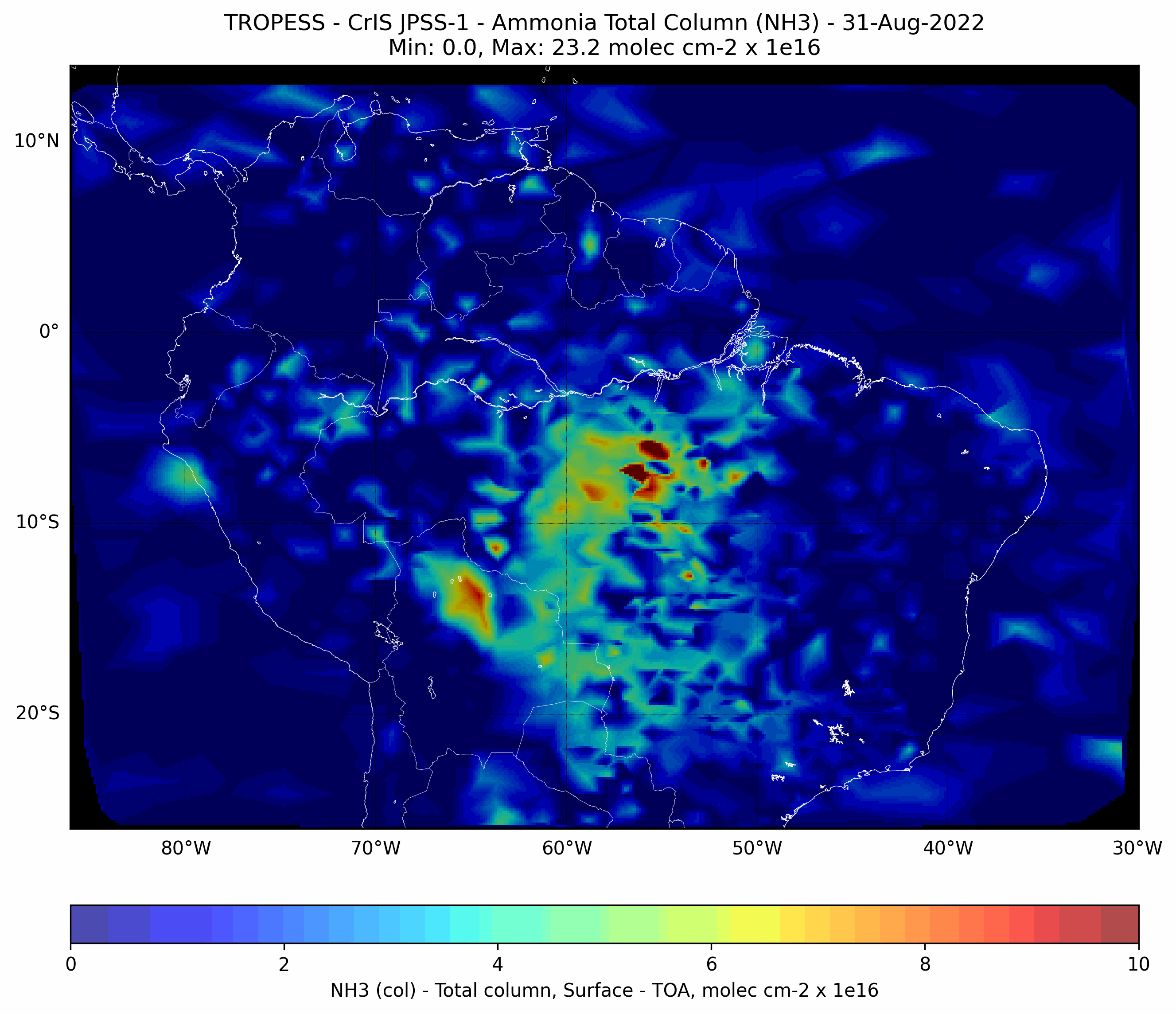
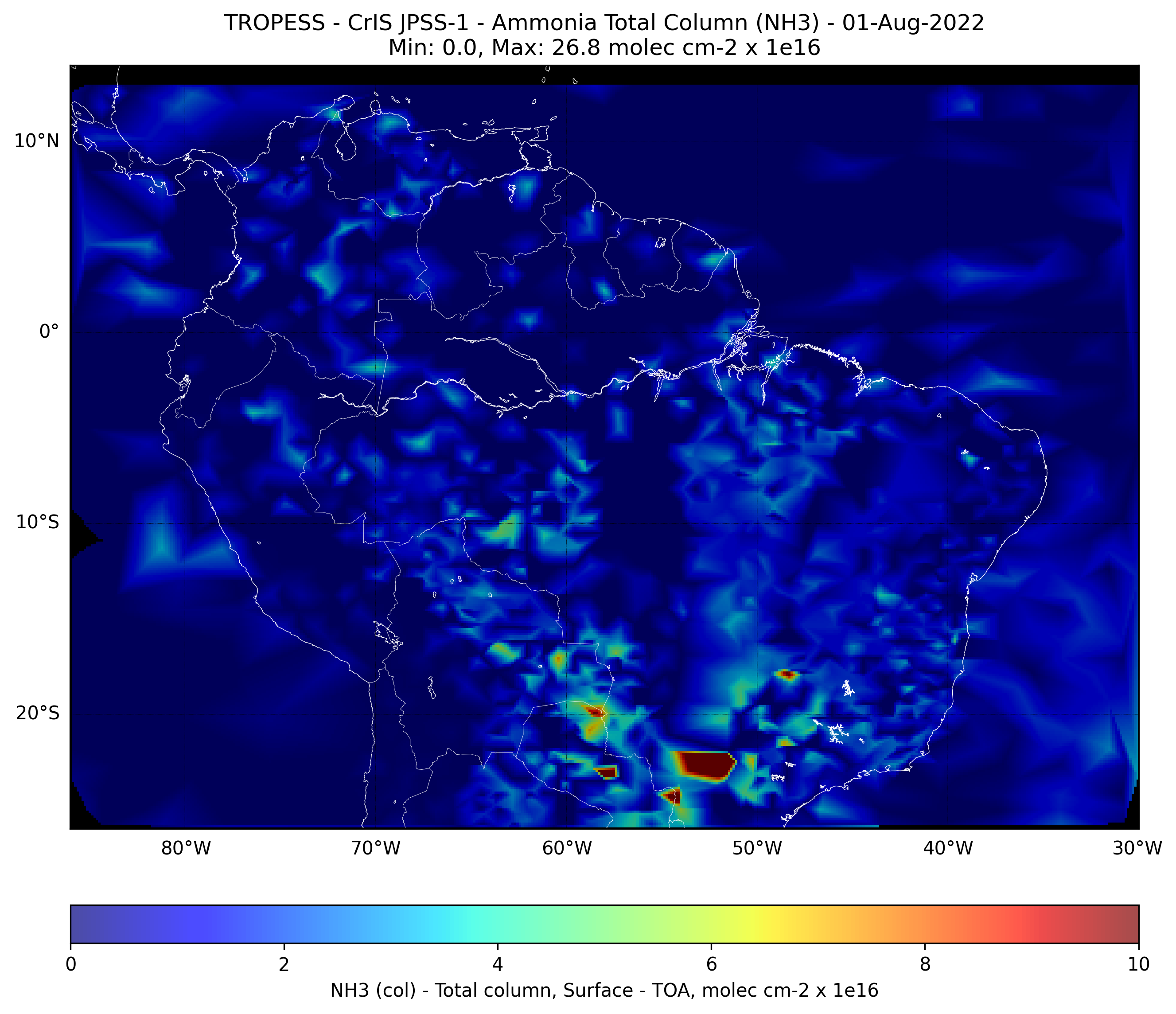
Brazilian Fires - Ammonia (NH3) - CrIS JPSS-1#
Total Column - gridded data (14km x 14km) plots#
This code plots the Ammonia (NH3) total column for each day from from 01-Aug-2022 to 31-Aug-2022.
import datetime
import contextlib
from pathlib import Path
import pandas as pd
import numpy as np
from netCDF4 import Dataset
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap
from scipy.interpolate import griddata
from scipy.spatial import cKDTree
import imageio.v2 as imageio
# Specify date range
start_date = datetime.date(2022, 8, 1)
end_date = datetime.date(2022, 8, 31)
# Loop through the date range and plot Carbon Monoxide for each day
for current_date in pd.date_range(start_date, end_date):
data_file_date = current_date.strftime("%Y%m%d")
# We may not have data for certain days
data_file_path = Path(f'./data/TROPESS_CrIS-JPSS1_L2_Summary_NH3_{data_file_date}_MUSES_R1p17_FS_F0p6.nc')
if not data_file_path.exists():
continue
# Open the netCDF file
dataset = Dataset(data_file_path.as_posix(), 'r')
# Read the data from your variables
latitude = dataset.variables['latitude'][:]
longitude = dataset.variables['longitude'][:]
x_col = dataset.variables['col'][:]
# scale to molec cm-2
x_col = x_col * dataset.variables['col'].for_molecules_per_cm2_multiply_by
x_col = x_col * 1e-16
dataset.close()
# Filter the data for South America only
latitude_max = 14.0
latitude_min = -26.0
longitude_max = -30.0
longitude_min = -86.0
south_america_only = np.where(
(latitude > latitude_min) & (latitude < latitude_max) &
(longitude > longitude_min) &(longitude < longitude_max)
)[0]
latitude = latitude[south_america_only]
longitude = longitude[south_america_only]
x_col = x_col[south_america_only]
# Calculate width and height
aspect_ratio = (longitude_max - longitude_min) / (latitude_max - latitude_min)
w = 3600; h = w / aspect_ratio
# Specify figure size (in inches)
dpi = 300;
plt.figure(figsize=(w / dpi, h / dpi))
# Create a basemap instance
m = Basemap(projection='cyl', resolution='i',
llcrnrlat=latitude_min, urcrnrlat=latitude_max, # set latitude limits to -56 and 24
llcrnrlon=longitude_min, urcrnrlon=longitude_max) # set longitude limits to -96 and -30
m.drawmapboundary(fill_color='black')
m.drawcoastlines(linewidth=0.3, color='white')
m.drawcountries(linewidth=0.2, color='white')
# Draw parallels (latitude lines) and meridians (longitude lines)
parallels = np.arange(-90., 91., 10.)
m.drawparallels(parallels, labels=[True,False,False,False], linewidth=0.3)
meridians = np.arange(-180., 181., 10.)
m.drawmeridians(meridians, labels=[False,False,False,True], linewidth=0.3)
# Get CrIS pixel size in degrees, pixel is 14 x 14 km, 1 degree is roughly 111.1 km
cris_pixel_size_deg = 14.0 / 111.1
# Get the grid for the interpolated values
grid_lat, grid_lon = np.mgrid[-26:13:cris_pixel_size_deg, -86:-29:cris_pixel_size_deg]
# Interpolate the data using griddata
grid_x_col = griddata((latitude, longitude), x_col, (grid_lat, grid_lon), method='linear', rescale=True)
# Find the distance to the nearest original point for each point in the interpolated grid
tree = cKDTree(np.vstack((latitude, longitude)).T)
dist, _ = tree.query(np.vstack((grid_lat.ravel(), grid_lon.ravel())).T)
# Reshape the distance array to have the same shape as the x_col grid
dist_grid = dist.reshape(grid_x_col.shape)
# Mask the interpolated values that are too far from any original point
max_distance_degrees = 3.0
grid_x_col[dist_grid > max_distance_degrees] = np.nan
# Plot the interpolated data using pcolormesh instead of scatter
sc = m.pcolormesh(grid_lon, grid_lat, grid_x_col, latlon=True, cmap='jet', alpha=0.7, vmin=0.0, vmax=10.0)
# Add a colorbar
cbar = m.colorbar(sc, location='bottom', pad="10%")
cbar.set_label('NH3 (col) - Total column, Surface - TOA, molec cm-2 x 1e16')
# set plot title
title_date = current_date.strftime("%d-%b-%Y")
title_min = np.min(x_col)
title_max = np.max(x_col)
plt.title(f'TROPESS - CrIS JPSS-1 - Ammonia Total Column (NH3) - {title_date}\nMin: {title_min:.01f}, Max: {title_max:.01f} molec cm-2 x 1e16')
# Save figure to PNG file
plt.savefig(f'./images/TROPESS_Brazilian-Fires_CrIS-JPSS-1_NH3_tc_{title_date}.png', dpi=dpi, bbox_inches='tight')
plt.close()
Now generate an animated GIF from the saved figures.
timelapse_start_date = start_date.strftime("%d-%b-%Y")
timelapse_end_date = end_date.strftime("%d-%b-%Y")
timelapse_file = f'./images/TROPESS_Brazilian-Fires_CrIS-JPSS-1_NH3_tc_Timelapse_{timelapse_start_date}_{timelapse_end_date}.gif'
# Path to the directory where the images were saved in the previous step
img_dir = Path('./images')
# Get all file names in the directory
files = sorted([img for img in img_dir.glob("TROPESS_Brazilian-Fires_CrIS-JPSS-1_NH3_tc*.png")])
# Read the images and add to a list
with contextlib.redirect_stdout(None):
images = []
for filename in files:
images.append(imageio.imread(filename))
# Save the images as an animated gif
# duration is the time spent on each image (in milliseconds)
imageio.mimsave(timelapse_file, images, duration=1000.0, loop=0)
First day#
Timelapse#